PepQuery WebSearch a peptide sequence against more than one billion MS/MS spectra indexed in PepQueryDB through the PepQuery web interface. |
PepQuery StandaloneSearch peptide sequences against PepQueryDB, public datasets in PRIDE, MassIVE, jPOSTrepo and iProX, or local MS/MS datasets. |
|---|
Welcome to PepQuery
PepQuery is a universal targeted peptide search engine for identifying or validating known and novel peptides of interest in any local or publicly available mass spectrometry-based proteomics datasets. The main functions of PepQuery include:
News
01/13/2023 PepQuery Web:
- Improved handling input format error.
12/21/2022 PepQuery Web:
- Fixed table randomly collapse issue.
- Improved showing long dataset string in "MS/MS dataset" drop-down list.
- Fixed a few other minor issues related to webpage.
11/23/2022 PepQuery Web:
- Improved error message handling.
- Fixed a bug when input sequence contains blank.
- Improved caching with error.
08/04/2022 PepQuery Web:
- Fixed a bug in setting DNA translation frame using the web interface.
08/02/2022 PepQuery 2.0.2:
- Fixed a bug in accessing PepQueryDB on Windows computers.
08/01/2022 PepQuery 2.0.1:
- Updated PepQueryDB access method. The previous version 2.0.0 is no longer able to access PepQueryDB.
- Updated two dependent Java libraries.
05/30/2022 PepQuery 2.0.0:
- Supported known peptide and protein identification;
- Supported PSM validation;
- Supported gene-centric peptide identification;
- Supported more MS/MS data formats;
- Supported directly searching proteomics data available in public data repositories including PRIDE, MassIVE, jPOSTrepo and iProX;
- Supported PSM validation from a USI;
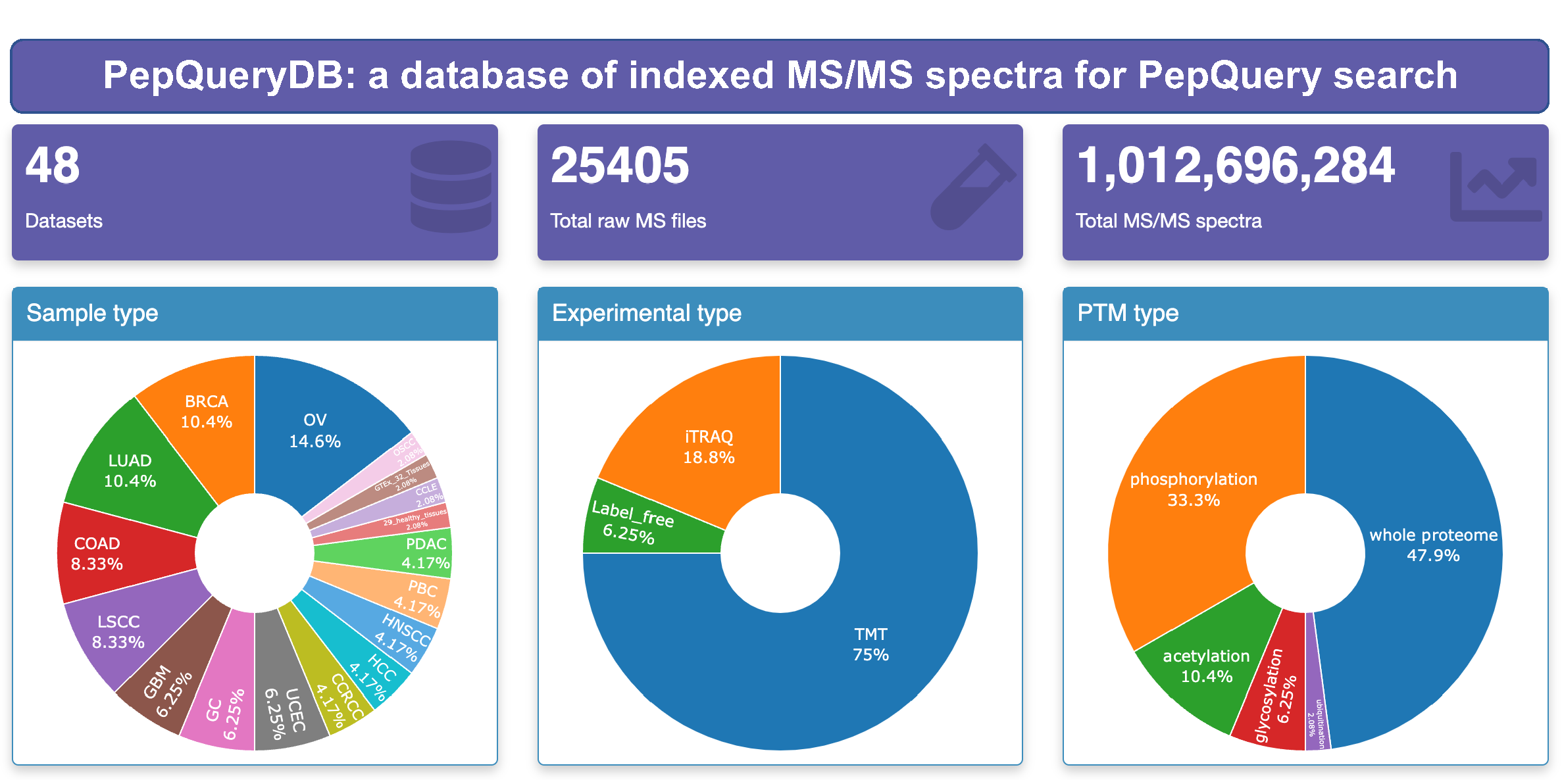
- New indexed MS/MS spectra database (PepQueryDB) which contains more than one billion spectra;
- All the public large scale CPTAC proteomics datasets available at PDC are included in PepQueryDB;
- Supported searching PepQueryDB in any computer with internet connection using the standalone version;
- Improved parameter setting: provided many predifined parameter settings for many different experiment workflows;
- Supported downloading protein reference database on the fly when run PepQuery;
- Supported modification setting for all the modifications in UniMod;
- Supported isotope error setting;
- Improved identification for phosphopeptide, acetylation (K) peptide and ubiquitination (K) peptide;
- Web server is updated to support the latest version;
- Improved spectrum annotation in the web server for modified peptides;
- Updated log4j to version 2.17.2;
- Various bug fixes and other improvements.
02/22/2021:
- Released PepQuery version 1.6.2.
02/01/2020:
- Released PepQuery version 1.4.1. In this version, we supported MHC ligand identification using immunopeptidomics data and fixed a few bugs.
07/28/2019:
- Released PepQuery version 1.2.0.
10/17/2018:
- Added a tool to support searching multiple datasets in a single run.
06/19/2018:
- Added more than 30 MS/MS datasets for the web server. This increased the total number of MS/MS spectra more than 0.5 billion;
- Updated PepQuery to output the best matches from the reference database searching for the spectra which matched to the input target peptides;
- Updated web server of PepQuery to present the best matched annotated spectra from the reference database searching.
01/05/2018:
- Updated protein reference database for dataset PASS00215_jurcat in the web server.
How to Cite
| Wen, Bo, Xiaojing Wang, and Bing Zhang. "PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations." Genome research 29.3 (2019): 485-493. |
|
| Wen, Bo, Kai Li, Yun Zhang, and Bing Zhang. "Cancer neoantigen prioritization through sensitive and reliable proteogenomics analysis." Nature communications 11.1 (2020): 1-14. |
|
| Wen, Bo, and Bing Zhang. "PepQuery2 democratizes public MS proteomics data for rapid peptide searching." Nature communications 14.1 (2023): 2213. |